BCI Kickstarter #07 : Building a P300 Speller: Translating Brainwaves into Letters
Welcome back to our BCI crash course! We've explored the foundations of BCIs, delved into the intricacies of brain signals, mastered the art of signal processing, and learned how to train intelligent algorithms to decode those signals. Now, we are ready to put all this knowledge into action by building a real-world BCI application: a P300 speller. P300 spellers are a groundbreaking technology that allows individuals with severe motor impairments to communicate by simply focusing their attention on letters on a screen. By harnessing the power of the P300 event-related potential, a brain response elicited by rare or surprising stimuli, these spellers open up a world of communication possibilities for those who might otherwise struggle to express themselves. In this blog, we will guide you through the step-by-step process of building a P300 speller using Python, MNE-Python, and scikit-learn. Get ready for a hands-on adventure in BCI development as we translate brainwaves into letters and words!

Step-by-Step Implementation: A Hands-on BCI Project
1. Loading the Dataset
Introducing the BNCI Horizon 2020 Dataset: A Rich Resource for P300 Speller Development
For this project, we'll use the BNCI Horizon 2020 dataset, a publicly available EEG dataset specifically designed for P300 speller research. This dataset offers several advantages:
- Large Sample Size: It includes recordings from a substantial number of participants, providing a diverse range of P300 responses.
- Standardized Paradigm: The dataset follows a standardized experimental protocol, ensuring consistency and comparability across recordings.
- Detailed Metadata: It provides comprehensive metadata, including information about stimulus presentation, participant responses, and electrode locations.
This dataset is well-suited for our P300 speller project because it provides high-quality EEG data recorded during a classic P300 speller paradigm, allowing us to focus on the core signal processing and machine learning steps involved in building a functional speller.
Loading the Data with MNE-Python: Accessing the Brainwave Symphony
To load the BNCI Horizon 2020 dataset using MNE-Python, you'll need to download the data files from the dataset's website (http://bnci-horizon-2020.eu/database/data-sets). Once you have the files, you can use the following code snippet to load a specific participant's data:
import mne
# Set the path to the dataset directory
data_path = '<path_to_dataset_directory>'
# Load the raw EEG data for a specific participant
raw = mne.io.read_raw_gdf(data_path + '/A01T.gdf', preload=True)
Replace <path_to_dataset_directory> with the actual path to the directory where you've stored the dataset files. This code loads the EEG data for participant "A01" during the training session ("T").
2. Data Preprocessing: Refining the EEG Signals for P300 Detection
Raw EEG data is often a mixture of brain signals, artifacts, and noise. Before we can effectively detect the P300 component, we need to clean up the data and isolate the relevant frequencies.
Channel Selection: Focusing on the P300's Neighborhood
The P300 component is typically most prominent over the central-parietal region of the scalp. Therefore, we'll select channels that capture activity from this area. Commonly used channels for P300 detection include:
- Cz: The electrode located at the vertex of the head, directly over the central sulcus.
- Pz: The electrode located over the parietal lobe, slightly posterior to Cz.
- Surrounding Electrodes: Additional electrodes surrounding Cz and Pz, such as CPz, FCz, and P3/P4, can also provide valuable information.
These electrodes are chosen because they tend to be most sensitive to the positive voltage deflection that characterizes the P300 response.
# Select the desired channels
channels = ['Cz', 'Pz', 'CPz', 'FCz', 'P3', 'P4']
# Create a new raw object with only the selected channels
raw_selected = raw.pick_channels(channels)
Filtering: Tuning into the P300 Frequency
The P300 component is a relatively slow brainwave, typically occurring in the frequency range of 0.1 Hz to 10 Hz. Filtering helps us remove unwanted frequencies outside this range, enhancing the signal-to-noise ratio for P300 detection.
We'll apply a band-pass filter to the selected EEG channels, using cutoff frequencies of 0.1 Hz and 10 Hz:
# Apply a band-pass filter from 0.1 Hz to 10 Hz
raw_filtered = raw_selected.filter(l_freq=0.1, h_freq=10)
This filter removes slow drifts (below 0.1 Hz) and high-frequency noise (above 10 Hz), allowing the P300 component to stand out more clearly.
Artifact Removal (Optional): Combating Unwanted Signals
Depending on the quality of the EEG data and the presence of artifacts, we might need to apply additional artifact removal techniques. Independent Component Analysis (ICA) is a powerful method for separating independent sources of activity in EEG recordings. If the BNCI Horizon 2020 dataset contains significant artifacts, we can use ICA to identify and remove components related to eye blinks, muscle activity, or other sources of interference.
3. Epoching and Averaging: Isolating the P300 Response
To capture the brain's response to specific stimuli, we'll create epochs, time-locked segments of EEG data centered around events of interest.
Defining Epochs: Capturing the P300 Time Window
We'll define epochs around both target stimuli (the letters the user is focusing on) and non-target stimuli (all other letters). The epoch time window should capture the P300 response, typically occurring between 300 and 500 milliseconds after the stimulus onset. We'll use a window of -200 ms to 800 ms to include a baseline period and capture the full P300 waveform.
# Define event IDs for target and non-target stimuli (refer to dataset documentation)
event_id = {'target': 1, 'non-target': 0}
# Set the epoch time window
tmin = -0.2 # 200 ms before stimulus onset
tmax = 0.8 # 800 ms after stimulus onset
# Create epochs
epochs = mne.Epochs(raw_filtered, events, event_id, tmin, tmax, baseline=(-0.2, 0), preload=True)
Baseline Correction: Removing Pre-Stimulus Bias
Baseline correction involves subtracting the average activity during the baseline period (-200 ms to 0 ms) from each epoch. This removes any pre-existing bias in the EEG signal, ensuring that the measured response is truly due to the stimulus.
Averaging Evoked Responses: Enhancing the P300 Signal
To enhance the P300 signal and reduce random noise, we'll average the epochs for target and non-target stimuli separately. This averaging process reveals the event-related potential (ERP), a characteristic waveform reflecting the brain's response to the stimulus.
# Average the epochs for target and non-target stimuli
evoked_target = epochs['target'].average()
evoked_non_target = epochs['non-target'].average()
4. Feature Extraction: Quantifying the P300
Selecting Features: Capturing the P300's Signature
The P300 component is characterized by a positive voltage deflection peaking around 300-500 ms after the stimulus onset. We'll select features that capture this signature:
- Peak Amplitude: The maximum amplitude of the P300 component.
- Mean Amplitude: The average amplitude within a specific time window around the P300 peak.
- Latency: The time it takes for the P300 component to reach its peak amplitude.
These features provide a quantitative representation of the P300 response, allowing us to train a classifier to distinguish between target and non-target stimuli.
Extracting Features: From Waveforms to Numbers
We can extract these features from the averaged evoked responses using MNE-Python's functions:
# Extract peak amplitude
peak_amplitude_target = evoked_target.get_data().max(axis=1)
peak_amplitude_non_target = evoked_non_target.get_data().max(axis=1)
# Extract mean amplitude within a time window (e.g., 300 ms to 500 ms)
mean_amplitude_target = evoked_target.crop(tmin=0.3, tmax=0.5).get_data().mean(axis=1)
mean_amplitude_non_target = evoked_non_target.crop(tmin=0.3, tmax=0.5).get_data().mean(axis=1)
# Extract latency of the P300 peak
latency_target = evoked_target.get_peak(tmin=0.3, tmax=0.5)[1]
latency_non_target = evoked_non_target.get_peak(tmin=0.3, tmax=0.5)[1]
5. Classification: Training the Brainwave Decoder
Choosing a Classifier: LDA for P300 Speller Decoding
Linear Discriminant Analysis (LDA) is a suitable classifier for P300 spellers due to its simplicity, efficiency, and ability to handle high-dimensional data. It seeks to find a linear combination of features that best separates the classes (target vs. non-target).
Training the Model: Learning from Brainwaves
We'll train the LDA classifier using the extracted features:
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
# Create an LDA object
lda = LinearDiscriminantAnalysis()
# Combine the features into a data matrix
X = np.vstack((peak_amplitude_target, peak_amplitude_non_target,
mean_amplitude_target, mean_amplitude_non_target,
latency_target, latency_non_target)).T
# Create a label vector (1 for target, 0 for non-target)
y = np.concatenate((np.ones(len(peak_amplitude_target)), np.zeros(len(peak_amplitude_non_target))))
# Train the LDA model
lda.fit(X, y)
Feature selection plays a crucial role here. By choosing features that effectively capture the P300 response, we improve the classifier's ability to distinguish between target and non-target stimuli.
6. Visualization: Validating Our Progress
Visualizing Preprocessed Data and P300 Responses
Visualizations help us understand the data and validate our preprocessing steps:
- Plot Averaged Epochs: Use evoked_target.plot() and evoked_non_target.plot() to visualize the average target and non-target epochs, confirming the presence of the P300 component in the target epochs.
- Topographical Plot: Use evoked_target.plot_topomap() to visualize the scalp distribution of the P300 component, ensuring it's most prominent over the expected central-parietal region.
Performance Evaluation: Assessing Speller Accuracy
Now that we've built our P300 speller, it's crucial to evaluate its performance. We need to assess how accurately it can distinguish between target and non-target stimuli, and consider practical factors that might influence its usability in real-world settings.
Cross-Validation: Ensuring Robustness and Generalizability
To obtain a reliable estimate of our speller's performance, we'll use k-fold cross-validation. This technique involves splitting the data into k folds, training the model on k-1 folds, and testing it on the remaining fold. Repeating this process k times, with each fold serving as the test set once, gives us a robust measure of the model's ability to generalize to unseen data.
from sklearn.model_selection import cross_val_score
# Perform 5-fold cross-validation
scores = cross_val_score(lda, X, y, cv=5)
# Print the average accuracy across the folds
print("Average accuracy: %0.2f" % scores.mean())
This code performs 5-fold cross-validation using our trained LDA classifier and prints the average accuracy across the folds.
Metrics for P300 Spellers: Beyond Accuracy
While accuracy is a key metric for P300 spellers, indicating the proportion of correctly classified stimuli, other metrics provide additional insights:
- Information Transfer Rate (ITR): Measures the speed of communication, taking into account the number of possible choices and the accuracy of selection. A higher ITR indicates a faster and more efficient speller.
Practical Considerations: Bridging the Gap to Real-World Use
Several practical factors can influence the performance and usability of P300 spellers:
- User Variability: P300 responses can vary significantly between individuals due to factors like age, attention, and neurological conditions. To address this, personalized calibration is crucial, where the speller is adjusted to each user's unique brain responses. Adaptive algorithms can also be employed to continuously adjust the speller based on the user's performance.
- Fatigue and Attention: Prolonged use can lead to fatigue and decreased attention, affecting P300 responses and speller accuracy. Strategies to mitigate this include incorporating breaks, using engaging stimuli, and employing algorithms that can detect and adapt to changes in user state.
- Training Duration: The amount of training a user receives can impact their proficiency with the speller. Sufficient training is essential for users to learn to control their P300 responses and achieve optimal performance.
Empowering Communication with P300 Spellers
We've successfully built a P300 speller, witnessing firsthand the power of EEG, signal processing, and machine learning to create a functional BCI application. These spellers hold immense potential as a communication tool, enabling individuals with severe motor impairments to express themselves, connect with others, and participate more fully in the world.
Further Reading and Resources
- Review article: Pan J et al. Advances in P300 brain-computer interface spellers: toward paradigm design and performance evaluation. Front Hum Neurosci. 2022 Dec 21;16:1077717. doi: 10.3389/fnhum.2022.1077717. PMID: 36618996; PMCID: PMC9810759.
- Dataset: BNCI Horizon 2020 P300 dataset: http://bnci-horizon-2020.eu/database/data-sets
- Tutorial: PyQt documentation for GUI development (optional): https://doc.qt.io/qtforpython/
Future Directions: Advancing P300 Speller Technology
The field of P300 speller development is constantly evolving. Emerging trends include:
- Deep Learning: Applying deep learning algorithms to improve P300 detection accuracy and robustness.
- Multimodal BCIs: Combining EEG with other brain imaging modalities (e.g., fNIRS) or physiological signals (e.g., eye tracking) to enhance speller performance.
- Hybrid Approaches: Integrating P300 spellers with other BCI paradigms (e.g., motor imagery) to create more flexible and versatile communication systems.
Next Stop: Motor Imagery BCIs
In the next blog post, we'll explore motor imagery BCIs, a fascinating paradigm where users control devices by simply imagining movements. We'll dive into the brain signals associated with motor imagery, learn how to extract features, and build a classifier to decode these intentions.

Capturing a biosignal is only the beginning. The real challenge starts once those tiny electrical fluctuations from your brain, heart, or muscles are recorded. What do they mean? How do we clean, interpret, and translate them into something both the machine and eventually we can understand? In this blog, we move beyond sensors to the invisible layer of algorithms and analysis that turns raw biosignal data into insight. From filtering and feature extraction to machine learning and real-time interpretation, this is how your body’s electrical language becomes readable.
Every heartbeat, every blink, every neural spark produces a complex trace of electrical or mechanical activity. These traces known collectively as biosignals are the raw currency of human-body intelligence. But in their raw form they’re noisy, dynamic, and difficult to interpret.
The transformation from raw sensor output to interpreted understanding is what we call biosignal processing. It’s the foundation of modern neuro- and bio-technology, enabling everything from wearable health devices to brain-computer interfaces (BCIs).
The Journey: From Raw Signal to Insight
When a biosignal sensor records, it captures a continuous stream of data—voltage fluctuations (in EEG, ECG, EMG), optical intensity changes, or pressure variations.
But that stream is messy. It includes baseline drift, motion artefacts, impedance shifts as electrodes dry, physiological artefacts (eye blinks, swallowing, jaw tension), and environmental noise (mains hum, electromagnetic interference).
Processing converts this noise-ridden stream into usable information, brain rhythms, cardiac cycles, muscle commands, or stress patterns.
Stage 1: Pre-processing — Cleaning the Signal
Before we can make sense of the body’s signals, we must remove the noise.
- Filtering: Band-pass filters (typically 0.5–45 Hz for EEG) remove slow drift and high-frequency interference; notch filters suppress 50/60 Hz mains hum.
- Artifact removal: Independent Component Analysis (ICA) and regression remain the most common methods for removing eye-blink (EOG) and muscle (EMG) artefacts, though hybrid and deep learning–based techniques are becoming more popular for automated denoising.
- Segmentation / epoching: Continuous biosignals are divided into stable time segments—beat-based for ECG or fixed/event-locked windows for EEG (e.g., 250 ms–1 s)—to capture temporal and spectral features more reliably.
- Normalization & baseline correction: Normalization rescales signal amplitudes across channels or subjects, while baseline correction removes constant offsets or drift to align signals to a common reference.
Think of this stage as cleaning a lens: if you don’t remove the smudges, everything you see through it will be distorted.
Stage 2: Feature Extraction — Finding the Patterns
Once the signal is clean, we quantify its characteristics, features that encode physiological or cognitive states.
Physiological Grounding
- EEG: Arises from synchronized postsynaptic currents in cortical pyramidal neurons.
- EMG: Records summed action potentials from contracting muscle fibers.
- ECG: Reflects rhythmic depolarization of cardiac pacemaker (SA node) cells.
Time-domain Features
Mean, variance, RMS, and zero-crossing rate quantify signal amplitude and variability over time. In EMG, Mean Absolute Value (MAV) and Waveform Length (WL) reflect overall muscle activation and fatigue progression.
Frequency & Spectral Features
The power of each EEG band tends to vary systematically across mental states.

Time–Frequency & Non-Linear Features
Wavelet transforms or Empirical Mode Decomposition capture transient events. Entropy- and fractal-based measures reveal complexity, useful for fatigue or cognitive-load studies.
Spatial Features
For multi-channel EEG, spatial filters such as Common Spatial Patterns (CSP) isolate task-specific cortical sources.
Stage 3: Classification & Machine Learning — Teaching Machines to Read the Body
After feature extraction, machine-learning models map those features to outcomes: focused vs fatigued, gesture A vs gesture B, normal vs arrhythmic.
- Classical ML: SVM, LDA, Random Forest , effective for curated features.
- Deep Learning: CNNs, LSTMs, Graph CNNs , learn directly from raw or minimally processed data.
- Transfer Learning: Improves cross-subject performance by adapting pretrained networks.
- Edge Inference: Deploying compact models (TinyML, quantized CNNs) on embedded hardware to achieve < 10 ms latency.
This is where raw physiology becomes actionable intelligence.
Interpreting Results — Making Sense of the Numbers
A robust pipeline delivers meaning, not just data:
- Detecting stress or fatigue for adaptive feedback.
- Translating EEG patterns into commands for prosthetics or interfaces.
- Monitoring ECG spectral shifts to flag early arrhythmias.
- Quantifying EMG coordination for rehabilitation or athletic optimization.
Performance hinges on accuracy, latency, robustness, and interpretability, especially when outcomes influence safety-critical systems.
Challenges and Future Directions
Technical: Inter-subject variability, electrode drift, real-world noise, and limited labeled datasets still constrain accuracy.
Ethical / Explainability: As algorithms mediate more decisions, transparency and consent are non-negotiable.
Multimodal Fusion: Combining EEG + EMG + ECG data improves reliability but raises synchronization and power-processing challenges.
Edge AI & Context Awareness: The next frontier is continuous, low-latency interpretation that adapts to user state and environment in real time.
Final Thought
Capturing a biosignal is only half the story. What truly powers next-gen neurotech and human-aware systems is turning that signal into sense. From electrodes and photodiodes to filters and neural nets, each link in this chain brings us closer to devices that don’t just measure humans; they understand them.

Every thought, heartbeat, and muscle twitch leaves behind a signal, but how do we actually capture them? In this blog post, we explore the sensors that make biosignal measurement possible, from EEG and ECG electrodes to optical and biochemical interfaces, and what it takes to turn those signals into meaningful data.
When we think of sensors, we often imagine cameras, microphones, or temperature gauges. But some of the most fascinating sensors aren’t designed to measure the world, they’re designed to measure you.
These are biosignal sensors: tiny, precise, and increasingly powerful tools that decode the electrical whispers of your brain, heart, and muscles. They're the hidden layer enabling brain-computer interfaces, wearables, neurofeedback systems, and next-gen health diagnostics.
But how do they actually work? And what makes one sensor better than another?
Let’s break it down, from scalp to circuit board.
First, a Quick Recap: What Are Biosignals?
Biosignals are the body’s internal signals, electrical, optical, or chemical , that reflect brain activity, heart function, muscle movement, and more. If you’ve read our earlier post on biosignal types, you’ll know they’re the raw material for everything from brain-computer interfaces to biometric wearables.
In this blog, we shift focus to the devices and sensors that make it possible to detect these signals in the real world, and what it takes to do it well.
The Devices That Listen In: Biosignal Sensor Types
.webp)
A Closer Look: How These Sensors Work
1. EEG / ECG / EMG – Electrical Sensors
These measure voltage fluctuations at the skin surface, caused by underlying bioelectric activity.
It’s like trying to hear a whisper in a thunderstorm; brain and muscle signals are tiny, and will get buried under noise unless the electrodes make solid contact and the amplifier filters aggressively.
There are two key electrode types:
- Wet electrodes: Use conductive gel or Saline for better signal quality. Still the gold standard in labs.
- Dry electrodes: More practical for wearables but prone to motion artifacts and noise (due to higher electrode resistance).
Signal acquisition often involves differential recording and requires high common-mode rejection ratios (CMRR) to suppress environmental noise.
Fun Fact: Even blinking your eyes generates an EMG signal that can overwhelm EEG data. That’s why artifact rejection algorithms are critical in EEG-based systems.
2. Optical Sensors (PPG, fNIRS)
These use light to infer blood flow or oxygenation levels:
- PPG: Emits light into the skin and measures reflection, pulsatile blood flow alters absorption.
- fNIRS: Uses near-infrared light to differentiate oxygenated vs. deoxygenated hemoglobin in the cortex.
Example: Emerging wearable fNIRS systems like Kernel Flow and OpenBCI Galea are making brain oxygenation measurement accessible outside labs.
3. Galvanic Skin Response / EDA – Emotion’s Electrical Signature
GSR (also called electrodermal activity) sensors detect subtle changes in skin conductance caused by sweat gland activity, a direct output of sympathetic nervous system arousal. When you're stressed or emotionally engaged, your skin becomes more conductive, and GSR sensors pick that up.
These sensors apply a small voltage across two points on the skin and track resistance over time. They're widely used in emotion tracking, stress monitoring, and psychological research due to their simplicity and responsiveness.
Together, these sensors form the foundation of modern biosignal acquisition — but capturing clean signals isn’t just about what you use, it’s about how you use it.
How Signal Quality Is Preserved
Measurement is just step one; capturing clean, interpretable signals involves:
- Analog Front End (AFE): Amplifies low signals while rejecting noise.
- ADC: Converts continuous analog signals into digital data.
- Signal Conditioning: Filters out drift, DC offset, 50/60Hz noise.
- Artifact Removal: Eye blinks, jaw clenches, muscle twitches.
Hardware platforms like TI’s ADS1299 and Analog Devices’ MAX30003 are commonly used in EEG and ECG acquisition systems.
New Frontiers in Biosignal Measurement
- Textile Sensors: Smart clothing with embedded electrodes for long-term monitoring.
- Biochemical Sensors: Detect metabolites like lactate, glucose, or cortisol in sweat or saliva.
- Multimodal Systems: Combining EEG + EMG + IMU + PPG in unified setups to boost accuracy.
A recent study involving transradial amputees demonstrated that combining EEG and EMG signals via a transfer learning model increased classification accuracy by 2.5–4.3% compared to EEG-only models.
Other multimodal fusion approaches, such as combining EMG and force myography (FMG), have shown classification improvements of over 10% compared to EMG alone.
Why Should You Care?
Because how we measure determines what we understand, and what we can build.
Whether it's a mental wellness wearable, a prosthetic limb that responds to thought, or a personalized neurofeedback app, it all begins with signal integrity. Bad data means bad decisions. Good signals? They unlock new frontiers.
Final Thought
We’re entering an era where technology doesn’t just respond to clicks, it responds to cognition, physiology, and intent.
Biosignal sensors are the bridge. Understanding them isn’t just for engineers; it’s essential for anyone shaping the future of human-aware tech.

In our previous blog, we explored how biosignals serve as the body's internal language—electrical, mechanical, and chemical messages that allow us to understand and interface with our physiology. Among these, electrical biosignals are particularly important for understanding how our nervous system, muscles, and heart function in real time. In this article, we’ll take a closer look at three of the most widely used electrical biosignals—EEG, ECG, and EMG—and their growing role in neurotechnology, diagnostics, performance tracking, and human-computer interaction. If you're new to the concept of biosignals, you might want to check out our introductory blog for a foundational overview.
"The body is a machine, and we must understand its currents if we are to understand its functions."-Émil du Bois-Reymond, pioneer in electrophysiology.
Life, though rare in the universe, leaves behind unmistakable footprints—biosignals. These signals not only confirm the presence of life but also narrate what a living being is doing, feeling, or thinking. As technology advances, we are learning to listen to these whispers of biology. Whether it’s improving health, enhancing performance, or building Brain-Computer Interfaces (BCIs), understanding biosignals is key.
Among the most studied biosignals are:
- Electroencephalogram (EEG) – from the brain
- Electrocardiogram (ECG) – from the heart
- Electromyogram (EMG) – from muscles
- Galvanic Skin Response (GSR) – from skin conductance
These signals are foundational for biosignal processing, real-time monitoring, and interfacing the human body with machines. In this article we look at some of these biosignals and some fascinating stories behind them.
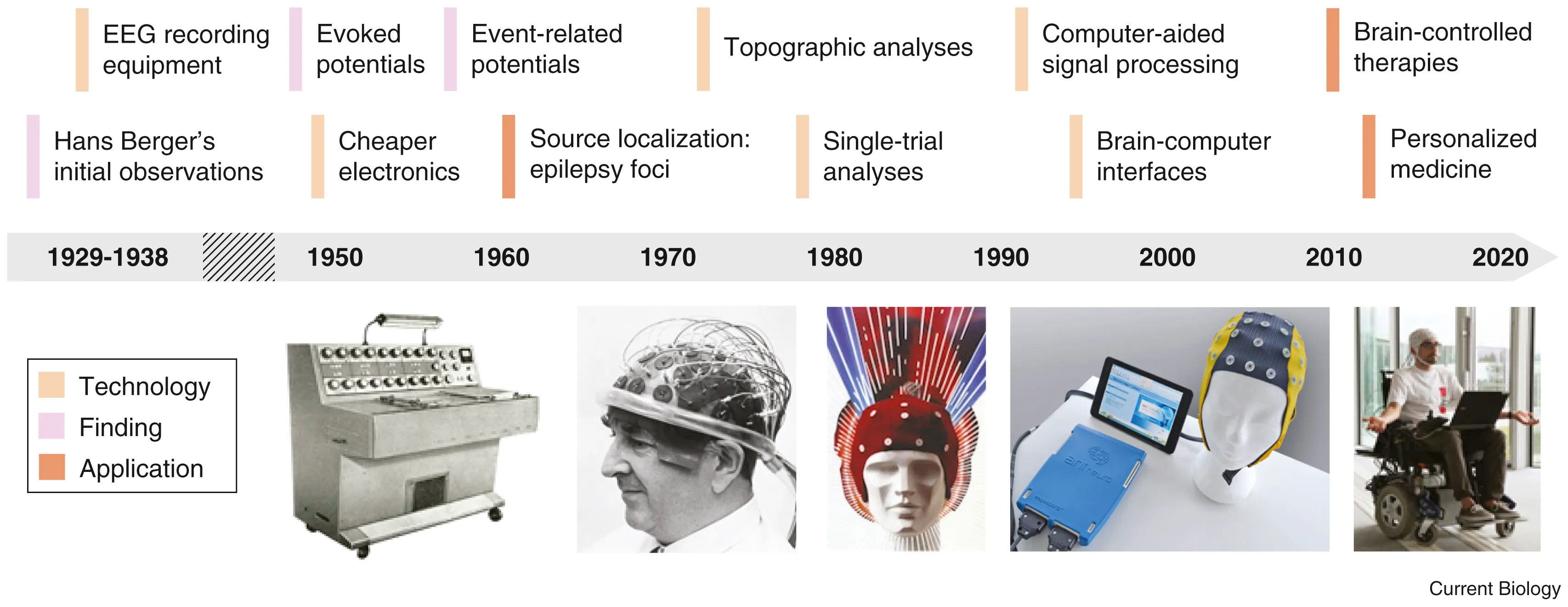
Electroencephalography (EEG): Listening to Brainwaves
In 1893, a 19 year old Hans Berger fell from a horse and had a near death experience. Little did he know that it would be a pivotal moment in the history of neurotechnology. The same day he received a telegram from his sister who was extremely concerned for him because she had a bad feeling. Hans Berger was convinced that this was due to the phenomenon of telepathy. After all, it was the age of radio waves, so why can’t there be “brain waves”? In his ensuing 30 year career telepathy was not established but in his pursuit, Berger became the first person to record brain waves.
When neurons fire together, they generate tiny electrical currents. These can be recorded using electrodes placed on the scalp (EEG), inside the skull (intracranial EEG), or directly on the brain (ElectroCorticogram). EEG signal processing is used not only to understand the brain’s rhythms but also in EEG-based BCI systems, allowing communication and control for people with paralysis. Event-Related Potentials (ERPs) and Local Field Potentials (LFPs) are specialized types of EEG signals that provide insights into how the brain responds to specific stimuli.
Electrocardiogram (ECG): The Rhythm of the Heart
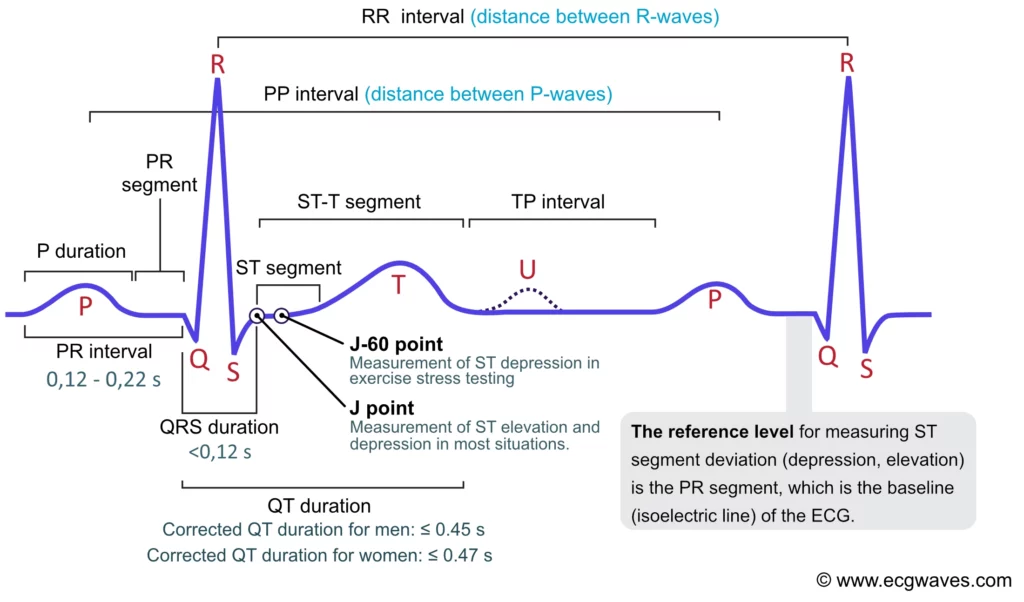
The heart has its own internal clock which produces tiny electrical signals every time it beats. Each heartbeat starts with a small electrical impulse made by a special part of the heart called the sinoatrial (SA) node. This impulse spreads through the heart muscle and makes it contract, first the upper (atria) and then lower chambers (ventricles) – that’s what pumps blood. This process produces voltage changes, which can be recorded via electrodes on the skin.
This gives rise to the classic PQRST waveform, with each component representing a specific part of the heart’s cycle. Modern wearables and medical devices use ECG signal analysis to monitor heart health in real time.
Fun fact: The waveform starts with “P” because Willem Einthoven left room for earlier letters—just in case future scientists discovered pre-P waves! So, thanks to a cautious scientist, we have the quirky naming system we still follow today.
Electromyography (EMG): The Language of Movement
When we perform any kind of movement - lifting our arm, kicking our leg, smiling, blinking or even breathing- our brain sends electrical signals to our muscles telling them to contract. When these neurons, known as motor neurons fire they release electrical impulses that travel to the muscle, causing it to contract. This electrical impulse—called a motor unit action potential (MUAP)—is what we see as an EMG signal. So, every time we move, we are generating an EMG signal!
Medical Applications
Medically, EMG is used for monitoring muscle fatigue especially in rehabilitation settings and muscle recovery post-injury or surgery. This helps clinicians measure progress and optimize therapy. EMG can distinguish between voluntary and involuntary movements, making it useful in diagnosing neuromuscular disorders, assessing stroke recovery, spinal cord injuries, and motor control dysfunctions.
Performance and Sports Science
In sports science, EMG can tell us muscle-activation timing and quantify force output of muscle groups. These are important factors to measure performance improvement in any sport. The number of motor units recruited and the synergy between muscle groups, helps us capture “mind-muscle connection” and muscle memory. Such things which were previously spoken off in a figurative manner can be scientifically measured and quantified using EMG. By tracking these parameters we get a window into movement efficiency and athletic performance. EMG is also used for biofeedback training, enabling individuals to consciously correct poor movement habits or retrain specific muscles
Beyond medicine and sports, EMG is used for gesture recognition in AR/VR and gaming, silent speech detection via facial EMG, and next-gen prosthetics and wearable exosuits that respond to the user’s muscle signals. EMG can be used in brain-computer interfaces (BCIs), helping paralyzed individuals control digital devices or communicate through subtle muscle activity. EMG bridges the gap between physiology, behavior, and technology—making it a critical tool in healthcare, performance optimization, and human-machine interaction.
As biosignal processing becomes more refined and neurotech devices more accessible, we are moving toward a world where our body speaks—and machines understand. Whether it’s detecting the subtlest brainwaves, tracking a racing heart, or interpreting muscle commands, biosignals are becoming the foundation of the next digital revolution. One where technology doesn’t just respond, but understands.

